揭示單細胞訊息系統的非線性特質
Reveal nonlinear properties of signaling systems in single cells
國立臺灣大學海洋所 謝志豪 特聘教授
中央研究院分生所 陳昇宏 副研究員
計畫執行期間:2023.1.1 – 2024.12.31
優良研究成果
➤共同重要成果突破Research results related to the National Taiwan University and Academia Sinica Innovative Joint Program after 2024 May
During this funding period of 國立臺灣大學與中央研究院創新性合作計畫, we have conducted the following projects and established an international collaboration with theoretical physicists in the Niels Bohr Institute, University of Copenhagen, Denmark –
Project 1: Plausible, robust biological oscillations through allelic buffering
Project 2: Fine-tuning p53 oscillatory dynamics for cell cycle-specific DNA damage response
Here, we provide a summary of these results as an update.
➤ 優良研究成果Published articles and manuscripts in preparation related to the project
Project1: Plausible, robust biological oscillations through allelic buffering
Biological oscillators are ubiquitous in nature, orchestrating biological functions in space and time via dedicated control of their oscillatory dynamics, which are primarily driven by feedback loops. However, these feedback loops involve multi-step biochemical reactions that inherently generate and amplify biochemical noise, raising the question of how biological oscillators, while repetitively activating stochastic biochemical reactions, maintain long-term robustness. To delineate the molecular mechanism underlying robust biological oscillators and characterizes the phenotypic consequences after crippling the robustness of oscillations, we take the oscillations of a cell-fate governor – p53 as an example. We showcase that biochemical noise causes stochastic formation of a high-level p53-Mdm2 inhibitory complex that suppresses MDM2 transcription, resulting in Mdm2 pulse skipping. Additionally, we demonstrate how a robust biological oscillator overcomes biochemical stochasticity by a novel buffering mechanism – allelic buffering, allowing for the precise control of cell fate.
This work is published Cell Systems 2024, Nov.
https://www.sciencedirect.com/science/article/pii/S2405471224002990
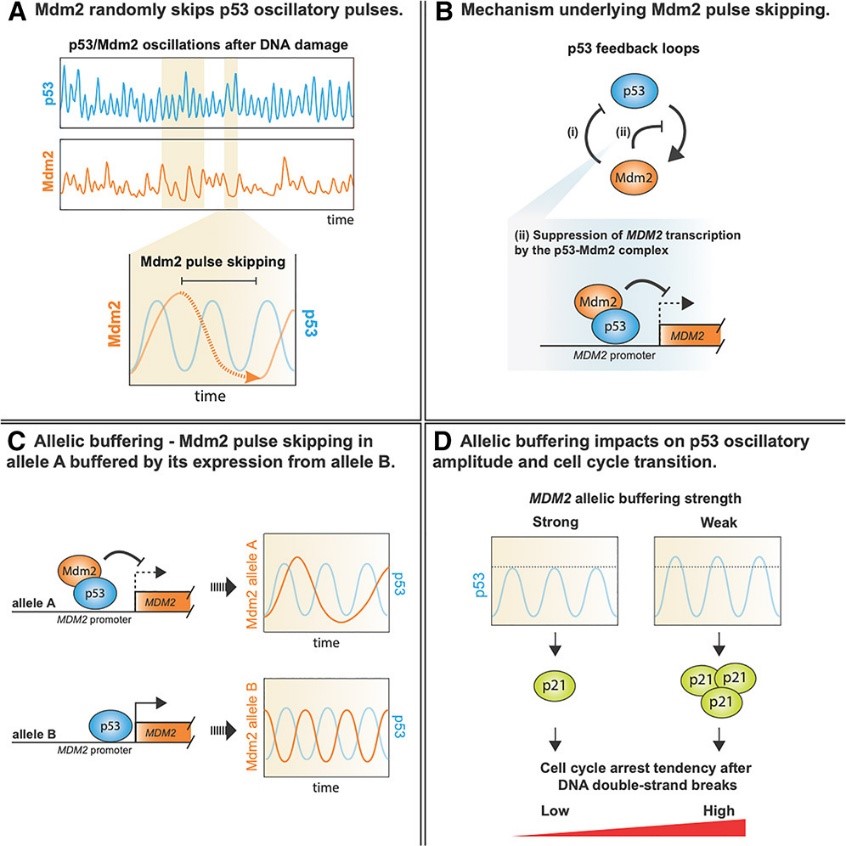
Project2: Fine-tuning p53 oscillatory dynamics for cell cycle-specific DNA damage response
Temporal dynamics of signaling molecules can encode biological information that governs physiological homeostasis and stress response. After DNA double strand breaks (DSBs), the tumor suppressor p53 can undergo long-term oscillations for cell cycle arrest. While the role of p53 in regulating cell cycle under genotoxic stress is well-established, it remains elusive how cell cycle position impacts on p53 oscillatory dynamics and how p53 oscillations, in turn, modulate DNA damage response in a cell cycle-specific manner. Using multiplexed p53 reporter cells, we found that p53 oscillator exhibits cell cycle-dependent dynamics – shorter periods and higher amplitudes of p53 oscillations in G1 phase compared to those in S/G2 phases. In addition, there are increases in p53’s period and amplitude during the cell cycle phase transition from G2 to 4N-G1. We further implemented the quantified, cell cycle-specific dynamical features into a mathematical model of the p53 oscillator to generate predictions about the possible underlying mechanisms driving this distinct oscillatory behavior. Taken together, our study not only showcases that p53 oscillator is biochemically fine-tuned in a cell cycle-specific manner, but also provides a framework for modulating p53 oscillatory dynamics to achieve cell cycle-specific DNA damage response.
This work is currently under manuscript preparation.